Abstract
Several genetic studies have shown an association between the 5-HT1A receptor gene (HTR1A) and major depressive disorder (MDD); however, results have been rather inconsistent. Moreover, to our knowledge, no association study on HTR1A and MDD in the Japanese population has been reported. Therefore, to evaluate the association between HTR1A and MDD, we conducted a case–control study of Japanese population samples with two single-nucleotide polymorphisms (SNPs), including rs6295 (C-1019G) in HTR1A. In addition, we conducted a meta-analysis of rs6295, which has been examined in other papers. Using one functional SNP (rs6295) and one tagging SNP (rs878567) selected with the HapMap database, we conducted a genetic association analysis of case–control samples (331 patients with MDD and 804 controls) in the Japanese population. Seven population-based association studies, including this study, met our criteria for the meta-analysis of rs6295. We found an association between rs878567 and Japanese MDD patients in the allele-wise analysis, but the significance of this association did not remain after Bonferroni's correction. We also did not detect any association between HTR1A and MDD in the allele/genotype-wise or haplotype-wise analysis. On the other hand, we detected an association between rs6295 and MDD in the meta-analysis (P(Z)=0.0327). In an explorative analysis, rs6295 was associated with Asian MDD patients after correction for multiple testing (P(Z)=0.0176), but not with Caucasian MDD patients (P(Z)=0.138). Our results suggest that HTR1A may not have a role in the pathophysiology of Japanese MDD patients. On the other hand, according to the meta-analysis, HTR1A was associated with MDD patients, especially in the Asian population.
Similar content being viewed by others
Introduction
Altered serotonergic neural transmission is hypothesized to be a susceptibility factor for major depressive disorder (MDD). The evidence for such an association is discussed in more detail in reviews.1, 2
Several genetic studies have shown an association between the serotonin 1A (5-HT1A) receptor gene (HTR1A) and MDD; however, results have been rather inconsistent. A recent meta-analysis showed no association between HTR1A and MDD.3 However, two very recent studies reported that rs6295 (C-1019G) in the promoter region of HTR1A, which regulates HTR1A transcription,4, 5 was associated with MDD in the Chinese population.6, 7 Moreover, to our knowledge, no association study of HTR1A and MDD in the Japanese population has been reported.
Therefore, we examined the association between HTR1A and MDD in the Japanese, using the recently recommended strategy of ‘gene-based’ association analysis.8 Moreover, we conducted an updated meta-analysis of rs6295, which has been intensively investigated in other studies.
Materials and methods
Subjects
The subjects in the association analysis were 331 patients with MDD (162 men and 169 women; mean age±s.d. 44.3±14.2 years) and 804 healthy controls (352 men and 452 women; mean age±s.d. 38.6±12.9 years). The patients were diagnosed according to the DSM-IV criteria with the consensus of at least two experienced psychiatrists on the basis of unstructured interviews and a review of medical records. In addition, at least 125 of the 331 MDD patients had been diagnosed according to the DSM-IV criteria with the consensus of at least two experienced psychiatrists on the basis of a review of medical records and assessment with the SIGH-D (Structured Interview Guide for Hamilton Rating Scale for Depression). All subjects were unrelated to each other, ethnically Japanese and lived in the central area of Japan. All healthy controls were also psychiatrically screened on the basis of unstructured interviews. None of them had severe medical complications, such as cirrhosis, renal failure, heart failure or other Axis-I disorders according to DSM-IV. No structured methods were used to assess psychiatric symptoms in the controls, which included hospital staff, their families and medical students.
The study was described to subjects and written informed consent was obtained from each. This study was approved by the ethics committee at Fujita Health University and Nagoya University School of Medicine.
SNP selection and linkage disequilibrium (LD) evaluation
We first consulted the HapMap database (release no. 23.a. phase2, March 2008, http://www.hapmap.org, population: Japanese Tokyo: minor allele frequencies (MAFs) of more than 0.05) and included three SNPs (rs6449693, rs878567 and rs1423691) covering HTR1A (5′-flanking regions including ∼1 kb from the initial exon and ∼2 kb downstream (3′) from the last exon: HapMap database contig number chr5: 63287418.63291774) (Figure 1). One tagging SNP was then selected with the criteria of an r2-threshold >0.8 in ‘pairwise-tagging-only’ mode using the ‘Tagger’ program (Paul de Bakker, http://www.broadinstitute.org/tagger-0) of the HAPLOVIEW software.9
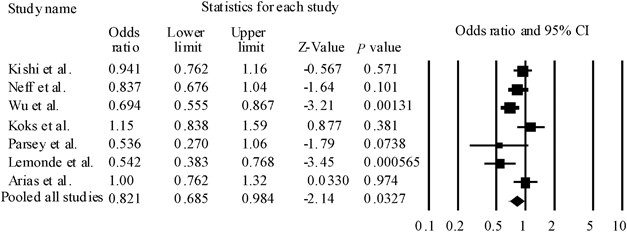
Forest plots of odds ratio (OR) with 95% confidence interval (95% CI) for rs6295. Results of all pooled studies are shown. We found significant heterogeneity among ORs (Q=16.2, df=6, P(Q)=0.0119). Therefore, we could calculate pooled ORs and P-values according to the DerSimonian and Laird random-effects model.
HTR1A has also been reported to have one biologically functional SNP (C-1019G: rs6295).4, 10, 11 rs6295 (C-1019G) in the promoter region regulates HTR1A transcription.4, 5 The C allele is a part of a 26 palindrome that connects transcription factors (Deaf-1, Hes1 and Hes5) by NUDR (nuclear deformed epidermal auto regulatory factor), whereas the G allele turns off repression by NUDR.4, 5 This would lead to elevated levels of 5-HT1A receptor in the presynaptic raphe nucleus in GG genotypes, compared with the CC genotype.4, 5 As no information about rs6295 was shown in the HapMap database, we included this SNP. These two SNPs were then used for the following association analysis.
SNP genotyping
We used TaqMan assays (Applied Biosystems, Foster City, CA, USA) for both SNPs. Detailed information, including primer sequences and reaction conditions, is available on request.
Statistical analysis
Case–control association study based on LD
Genotype deviation from the Hardy–Weinberg equilibrium (HWE) was evaluated by χ2 test (SAS/Genetics, release 8.2, SAS Japan, Tokyo, Japan).
Marker-trait association analysis was used to evaluate allele- and genotype-wise association with the χ2 test (SAS/Genetics, release 8.2, SAS Japan), and haplotype-wise association analysis was conducted with a likelihood ratio test using the COCAPHASE2.403 program.12 Power calculation was performed using the Genetic Power Calculator.13 Bonferroni's correction was used to control inflation of the type I error rate. The significance level for all statistical tests was 0.05.
Meta-analysis
To identify studies eligible for the meta-analysis, we searched PubMed citations through March 2009 using the terms ‘HTR1A,’ ‘serotonin 1A receptor gene,’ ‘major depressive disorder’ and ‘MDD’ as key words.
As criteria selected for eligible studies, we referred to the study by López-León et al.3 In summary, eligible studies had to meet all of the following criteria: (1) be published in peer-reviewed journal, (2) have cases not selected by questionnaires assessing symptoms of depression, (3) contain independent data, (4) have distribution of genotypes in the control population that was in the HWE, (5) be case–control association studies investigating one or more of the three polymorphisms, (6) have MDD patients diagnosed according to ICD and DSM and (7) use healthy individuals as controls in case–control studies.
Cochran's χ2-based Q-statistic test was applied to assess between-study heterogeneity. The significance of the pooled odds ratio (OR) was determined using a Z-test. Overall, ORs and their 95% confidence intervals (95% CIs) were estimated under both the Mantel–Haenszel14 fixed-effects and DerSimonian–Laird15 random-effects models. The random-effects model is more conservative than is the fixed-effects model and produces a wider CI. When there is no evidence of heterogeneity, the random-effects model will yield similar results to the fixed-effects model. Therefore, if it is confirmed that there was no heterogeneity, we could calculate pooled ORs and P-values according to the Mantel–Haenszel fixed-effects model. If there was evidence of heterogeneity, we could calculate pooled ORs and P-values according to the DerSimonian and Laird random-effects model. Publication bias was evaluated using a funnel plot asymmetry with Egger's test. The statistical significance was set at 0.05. All data were analyzed using Comprehensive Meta Analysis (Version 2.0). More detailed information about the meta-analysis method is given in our previous papers. Bonferroni's correction was used to control inflation of the type I error rate. The significance level for all statistical tests was 0.05.
Results
The LD from rs6449693, rs878567 and rs1423691 was tight, according to the HapMap database samples (r2=1.00). However, the LD structure of rs6295 (functional SNP) and rs878567 (tagging SNP) in our control samples was not tight (r2=0.160). Genotype frequencies of all SNPs were in the HWE. We found an association between rs878567 and Japanese MDD patients in the allele-wise analysis (P=0.0448), but this significance did not remain after Bonferroni's correction (P=0.0896). We also did not detect any association between HTR1A and MDD in the allele/genotype-wise (Table 1) or in the haplotype-wise analysis (P=0.126).
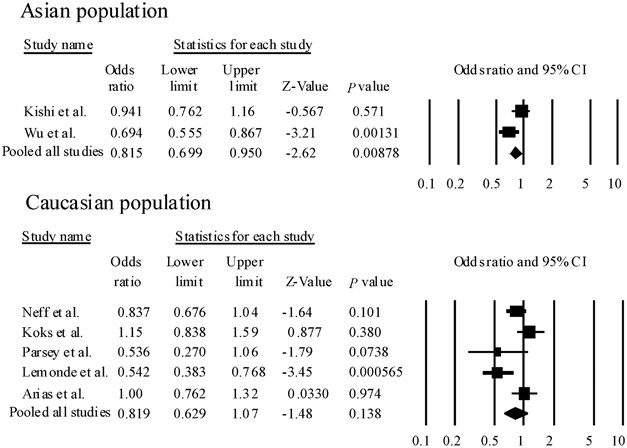
In the meta-analysis, seven population-based association studies, including this study, met our criteria for rs62954, 6, 16, 17, 18, 19 (Table 2). We found significant heterogeneity among ORs (Q=16.2, df=6, P(Q)=0.0119). The pooled OR derived from all studies comprising 1658 patients and 2046 control subjects indicated a significant association (random model: OR=0.821, 95% CI=0.695–984, P(Z)=0.0327) (Table 2) (Figure 1). Next, to limit ethnic heterogeneity, we included an explorative analysis with either Caucasian or Asian samples. We did not observe significant heterogeneity among ORs in the Asian population (Q=4.30, df=1, P(Q)=0.133). However, we found significant heterogeneity among ORs in the Caucasian population (Q=13.1, df=4, P(Q)=0.0108). We detected a significant association between rs6295 and MDD in the Asian population (fixed model: pooled OR=0.815, 95% CI=0.699–0.950, P(Z)=0.00878) (Figure 2). Moreover, these associations were still significant after correction for multiple testing (fixed model: P(Z)=0.0176) (Figure 2). In addition, rs6295 was not associated with MDD in the Caucasian population (random model: OR=0.819, 95% CI=0.629–1.066, P(Z)=0.138) (Figure 1). No publication bias was found (t=0.685, P=0.524).
Forest plots of odds ratio (OR) with 95% confidence interval (95% CI) for rs6295. Results of subgroup analysis are shown. We did not observe significant heterogeneity among ORs in the Asian population (Q=4.30, df=1, P(Q)=0.133). Therefore, we could calculate pooled ORs and P-values according to the Mantel–Haenszel fixed-effects model. However, we found significant heterogeneity among ORs in the Caucasian population (Q=13.1, df=4, P(Q)=0.0108). Therefore, we could calculate pooled ORs and P-values according to the DerSimonian and Laird random-effects model.
Discussion
We detected an association between rs6295 and MDD in the meta-analysis. However, because significant heterogeneity among ORs was found (P=0.0119), we carried out an explorative analysis with either Caucasian or Asian samples. We detected a significant association between rs6295 and MDD in the Asian population, but did not observe significant heterogeneity among ORs (P=0.133). On the other hand, we observed significant heterogeneity among ORs in the Caucasian population (P=0.0108). The heterogeneity may result from (1) different ancestries (Asian population versus Caucasian population), (2) incomplete genotyping rates or genotyping error rates in different studies. Although the studies by Arias et al.16 and Koks et al.18 reported that the major allele was ‘G’, other studies showed that the major allele was ‘C’. We contacted the corresponding authors with regard to the genotype data in these studies. They and Kato et al.20 informed us that there were no genotyping errors. (3) The overall sample size included in the meta-analysis was relatively small (1658 patients and 2046 control subjects).21 To overcome these limitations, a replication study using larger samples or samples of other populations will be required for conclusive results.
In our case–control study, no association was found. Although the Japanese and Chinese populations are both classified as Asian populations, rs6295 was associated with Chinese MDD patients,6, 7 but not with Japanese MDD patients. Several reasons may explain these inconsistent findings, including allelic heterogeneity, true variation in disease association between populations, modifying genetic and/or environmental factors and statistically underpowered small sample sizes. In rs6295, the MAFs in MDD in the Japanese population (0.246) seem to be smaller compared with those in the Chinese population (0.303).6 The MAFs in Japanese controls were similar to those in Chinese controls (0.231).6 Although classified in the same Asian population, the susceptibility genes for MDD might be not common. Recent evidence supports this hypothesis. The serotonin 2A gene was associated with MDD in the Korean population,22 but not in Japanese population.23 rs6295 (C-1019G) in the promoter region regulates HTR1A transcription.4, 5 The C allele is part of a 26 palindrome that connects transcription factors (Deaf-1, Hes1 and Hes5) by NUDR, whereas the G allele turns off repression by NUDR.4, 5 This would lead to elevated levels of 5-HT1A receptor in the presynaptic raphe nucleus in GG genotypes compared with the CC genotype.4, 5 In several studies, these variants were associated with responses including the antidepressant response in MDD.24, 25, 26, 27, 28, 29 Recently, Kato and Serretti30 reported a meta-analysis between rs6295 and antidepressant efficacy in MDD. The authors showed that the pooled OR derived from six studies comprising 893 patients was not significant.30 The pooled OR of studies in Caucasian populations only was also not significant.30 However, the pooled OR of the studies in Asian studies only did show a significant association.30 Therefore, the authors suggested that the rs6295 genotype may be a predictor of antidepressant treatment response in MDD in the Asian population.30
A few points of caution should be mentioned with respect to our results. First, our sample sizes were small. In the power analysis, we obtained power of more than 80% for the detection of association when we set the genotype relative risk at 1.33–1.51 in MDD for HTR1A, under a multiplicative model of inheritance. As our samples were small, type II errors are possible in the results of these statistical association analyses. Second, we did not perform a mutation scan of HTR1A. As we consider it to be difficult to evaluate the association of such extremely rare variants from the viewpoint of statistical power, a replication study using a larger sample is required for conclusive results. Finally, our subjects did not undergo structured interviews. MDD patients who are not diagnosed by structured interview may develop bipolar disorder in the future.31 However, in this study, patients were carefully diagnosed according to the DSM-IV criteria with the consensus of at least two experienced psychiatrists on the basis of a review of medical records. In addition, when we found a misdiagnosis in a patient, we promptly excluded the misdiagnosed case in consideration of the precision of our sample. Detailed information on our samples was provided in previous papers.32, 33, 34, 35
In conclusion, our results suggest that HTR1A may not have a role in the pathophysiology of Japanese MDD patients. On the other hand, according to the meta-analysis, HTR1A was associated with MDD patients, especially in the Asian population. As our sample and the overall sample size included in the meta-analysis were relatively small, statistical errors are possible in the results of these statistical association analyses. To overcome these limitations, a replication study using a larger sample may be required for conclusive results.
References
Kato, T. Molecular genetics of bipolar disorder and depression. Psychiatry Clin Neurosci 61, 3–19 (2007).
Levinson, D. F. The genetics of depression: a review. Biol Psychiatry 60, 84–92 (2006).
López-León, S., Janssens, A. C., Gonzalez-Zuloeta Ladd, A. M., Del-Favero, J., Claes, S. J., Oostra, B. A. et al. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry 13, 772–785 (2008).
Lemonde, S., Turecki, G., Bakish, D., Du, L., Hrdina, P. D., Bown, C. D. et al. Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J Neurosci 23, 8788–8799 (2003).
Le Francois, B., Czesak, M., Steubl, D., Albert, P. R. Transcriptional regulation at a HTR1A polymorphism associated with mental illness. Neuropharmacology 55, 977–985 (2008).
Wu, Y., Xu, Y., Sun, Y., Wang, Y. F., Li, X., Lang, X. E. et al. Association between the serotonin 1A receptor C(-1019)G polymorphism and major depressive disorder in the northern Han ethnic group in China. Chin Med J 121, 874–876 (2008).
Zhang, K., Xu, Q., Xu, Y., Yang, H., Luo, J., Sun, Y. et al. The combined effects of the 5-HTTLPR and 5-HTR1A genes modulates the relationship between negative life events and major depressive disorder in a Chinese population. J Affect Disord 114, 224–231 (2009).
Neale, B. M. & Sham, P. C. The future of association studies: gene-based analysis and replication. Am J Hum Genet 75, 353–362 (2004).
Barrett, J. C., Fry, B., Maller, J. & Daly, M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England) 21, 263–265 (2005).
Albert, P. R., Lembo, P., Storring, J. M., Charest, A. & Saucier, C. The 5-HT1A receptor: signaling, desensitization, and gene transcription. Neuropsychopharmacology 14, 19–25 (1996).
Albert, P. R. & Lemonde, S. 5-HT1A receptors, gene repression, and depression: guilt by association. Neuroscientist 10, 575–593 (2004).
Dudbridge, F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 25, 115–121 (2003).
Purcell, S., Cherny, S. S. & Sham, P. C. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics (Oxford, England) 19, 149–150 (2003).
Mantel, N. & Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–748 (1959).
DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–188 (1986).
Arias, B., Arranz, M. J., Gasto, C., Catalan, R., Pintor, L., Gutierrez, B. et al. Analysis of structural polymorphisms and C-1018G promoter variant of the 5-HT(1A) receptor gene as putative risk factors in major depression. Mol Psychiatry 7, 930–932 (2002).
Neff, C. D., Abkevich, V., Packer, J. C., Chen, Y., Potter, J., Riley, R. et al. Evidence for HTR1A and LHPP as interacting genetic risk factors in major depression. Mol Psychiatry 14, 621–630 (2008).
Koks, S., Nikopensius, T., Koido, K., Maron, E., Altmae, S., Heinaste, E. et al. Analysis of SNP profiles in patients with major depressive disorder. Int J Neuropsychopharmacol/Official Scientific Journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 9, 167–174 (2006).
Parsey, R. V., Oquendo, M. A., Ogden, R. T., Olvet, D. M., Simpson, N., Huang, Y. Y. et al. Altered serotonin 1A binding in major depression: a [carbonyl-C-11]WAY100635 positron emission tomography study. Biol Psychiatry 59, 106–113 (2006).
Kato, M., Fukuda, T., Wakeno, M., Okugawa, G., Takekita, Y., Watanabe, S. et al. Effect of 5-HT1A gene polymorphisms on antidepressant response in major depressive disorder. Am J Med Genet B Neuropsychiatr Genet 150B, 115–123 (2009).
Shi, J., Badner, J. A., Gershon, E. S. & Liu, C. Allelic association of G72/G30 with schizophrenia and bipolar disorder: a comprehensive meta-analysis. Schizophrenia Res 98, 89–97 (2008).
Choi, M. J., Lee, H. J., Lee, H. J., Ham, B. J., Cha, J. H., Ryu, S. H. et al. Association between major depressive disorder and the -1438A/G polymorphism of the serotonin 2A receptor gene. Neuropsychobiology 49, 38–41 (2004).
Kishi, T., Kitajima, T., Tsunoka, T., Ikeda, M., Yamanouchi, Y., Kinoshita, Y. et al. Genetic association analysis of serotonin 2A receptor gene (HTR2A) with bipolar disorder and major depressive disorder in the Japanese population. Neurosci Res 64, 231–234 (2009).
Serretti, A., Artioli, P., Lorenzi, C., Pirovano, A., Tubazio, V. & Zanardi, R. The C(-1019)G polymorphism of the 5-HT1A gene promoter and antidepressant response in mood disorders: preliminary findings. Int J Neuropsychopharmacology/Official Scientific Journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 7, 453–460 (2004).
Lemonde, S., Du, L., Bakish, D., Hrdina, P. & Albert, P. R. Association of the C(-1019)G 5-HT1A functional promoter polymorphism with antidepressant response. Int J Neuropsychopharmacology/Official Scientific Journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 7, 501–506 (2004).
Arias, B., Catalan, R., Gasto, C., Gutierrez, B. & Fananas, L. Evidence for a combined genetic effect of the 5-HT(1A) receptor and serotonin transporter genes in the clinical outcome of major depressive patients treated with citalopram. J Psychopharmacol (Oxford, England) 19, 166–172 (2005).
Parsey, R. V., Olvet, D. M., Oquendo, M. A., Huang, Y. Y., Ogden, R. T. & Mann, J. J. Higher 5-HT1A receptor binding potential during a major depressive episode predicts poor treatment response: preliminary data from a naturalistic study. Neuropsychopharmacology 31, 1745–1749 (2006).
Hong, C. J., Chen, T. J., Yu, Y. W. & Tsai, S. J. Response to fluoxetine and serotonin 1A receptor (C-1019G) polymorphism in Taiwan Chinese major depressive disorder. Pharmacogenomics J 6, 27–33 (2006).
Yu, Y. W., Tsai, S. J., Liou, Y. J., Hong, C. J. & Chen, T. J. Association study of two serotonin 1A receptor gene polymorphisms and fluoxetine treatment response in Chinese major depressive disorders. Eur Neuropsychopharmacol 16, 498–503 (2006).
Kato, M. & Serretti, A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry (2008) (in press).
Bowden, C. L. Strategies to reduce misdiagnosis of bipolar depression. Psychiatr Serv (Washington, DC) 52, 51–55 (2001).
Kishi, T., Ikeda, M., Kitajima, T., Suzuki, T., Yamanouchi, Y., Kinoshita, Y. et al. No association between prostate apoptosis response 4 gene (PAWR) in schizophrenia and mood disorders in a Japanese population. Am J Med Genet B Neuropsychiatr Genet 147B, 531–534 (2008).
Kishi, T., Kitajima, T., Ikeda, M., Yamanouchi, Y., Kinoshita, Y., Kawashima, K. et al. Association study of clock gene (CLOCK) and schizophrenia and mood disorders in the Japanese population. Eur Arch Psychiatry Clin Neurosci 259, 293–297 (2009).
Kishi, T., Kitajima, T., Ikeda, M., Yamanouchi, Y., Kinoshita, Y., Kawashima, K. et al. Association analysis of nuclear receptor Rev-erb alpha gene (NR1D1) with mood disorders in the Japanese population. Neurosci Res 62, 211–215 (2008).
Kishi, T., Kitajima, T., Tsunoka, T., Ikeda, M., Yamanouchi, Y., Kinoshita, Y. et al. Genetic association analysis of serotonin 2A receptor gene (HTR2A) with bipolar disorder and major depressive disorder in the Japanese population. Neurosci Res 64, 231–234 (2009).
Acknowledgements
We thank Ms M Miyata and Ms S Ishihara for their technical support. This work was supported in part by research grants from the Ministry of Education, Culture, Sports, Science and Technology, the Ministry of Health, Labor and Welfare, and the Japan Health Sciences Foundation (Research on Health Sciences focusing on Drug Innovation).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kishi, T., Tsunoka, T., Ikeda, M. et al. Serotonin 1A receptor gene and major depressive disorder: an association study and meta-analysis. J Hum Genet 54, 629–633 (2009). https://doi.org/10.1038/jhg.2009.84
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.84
Keywords
This article is cited by
-
The effect of negative life events on college students’ depression: the mediating role of internet addiction and the moderating role of 5-HTT1A gene rs6449693 polymorphism
Current Psychology (2023)
-
Dissecting diagnostic heterogeneity in depression by integrating neuroimaging and genetics
Neuropsychopharmacology (2021)
-
Serotonergic gene polymorphisms (5-HTTLPR, 5HTR1A, 5HTR2A), and population differences in aggression: traditional (Hadza and Datoga) and industrial (Russians) populations compared
Journal of Physiological Anthropology (2018)
-
The role of 5-HT receptors in depression
Molecular Brain (2017)
-
5-HT1A Autoreceptors in the Dorsal Raphe Nucleus Convey Vulnerability to Compulsive Cocaine Seeking
Neuropsychopharmacology (2016)