Abstract
Background: Previous studies have shown that individuals with schizophrenia have a greater risk for psoriasis than a typical person. This suggests that there might be a shared genetic etiology between the 2 conditions. We aimed to characterize the potential shared genetic susceptibility between schizophrenia and psoriasis using genome-wide marker genotype data.
Methods: We obtained genetic data on individuals with psoriasis, schizophrenia and control individuals. We applied a marker-based coheritability estimation procedure, polygenic score analysis, a gene set enrichment test and a least absolute shrinkage and selection operator regression model to estimate the potential shared genetic etiology between the 2 diseases. We validated the results in independent schizophrenia and psoriasis cohorts from Singapore.
Results: We included 1139 individuals with psoriasis, 744 with schizophrenia and 1678 controls in our analysis, and we validated the results in independent cohorts, including 441 individuals with psoriasis (and 2420 controls) and 1630 with schizophrenia (and 1860 controls). We estimated that a large fraction of schizophrenia and psoriasis risk could be attributed to common variants (h2SNP = 29% ± 5.0%, p = 2.00 × 10−8), with a coheritability estimate between the traits of 21%. We identified 5 variants within the human leukocyte antigen (HLA) gene region, which were most likely to be associated with both diseases and collectively conferred a significant risk effect (odds ratio of highest risk quartile = 6.03, p < 2.00 × 10−16). We discovered that variants contributing most to the shared heritable component between psoriasis and schizophrenia were enriched in antigen processing and cell endoplasmic reticulum.
Limitations: Our sample size was relatively small. The findings of 5 HLA gene variants were complicated by the complex structure in the HLA region.
Conclusion: We found evidence for a shared genetic etiology between schizophrenia and psoriasis. The mechanism for this shared genetic basis likely involves immune and calcium signalling pathways.
Introduction
Schizophrenia and psoriasis are common complex disorders with large immune components.1,2 Schizophrenia has been shown to be associated with numerous autoimmune diseases, and psoriasis is a common immune-mediated inflammatory skin disease. Both result in a heavy societal and public health burden. Schizophrenia and psoriasis both have an established genetic component,3,4 and are relatively common in the Han Chinese population (approximate prevalence of 1% and 0.5%, respectively4,5). Cases of clinical comorbidity involving both schizophrenia and psoriasis have been reported.6 Epidemiological studies of the Taiwan Chinese population have shown that psoriasis develops in about 0.87% of patients with schizophrenia, which is a higher proportion than in the general population.7,8 In addition, a study involving individuals in the Danish nationwide registry suggests that the incidence of psoriasis is nearly twice as high in patients with schizophrenia than the general population. Moreover, they found that the incidence of autoimmune disorders in patients with schizophrenia or individuals with a family history of schizophrenia is increased. It is estimated that autoimmune diseases develop in about 3.6% of individuals with schizophrenia and that about 3.1% of people with autoimmune diseases have a family history of schizophrenia.9,10 In light of these findings, it is quite likely that comorbidity of schizophrenia and psoriasis arises as a result of a shared autoimmune dysfunction.1,2,11
Previous results from our genome-wide association studies (GWAS) conducted independently in Han Chinese schizophrenia and psoriasis cohorts suggested that a shared susceptibility to these disorders may stem from genetic variants on chromosome 6, as it was found to be associated with both disorders. It is noteworthy that the human leukocyte antigen (HLA) gene has been implicated in many immune disorders.3,4 In addition, other researchers have identified variants in the CSMD1 gene as a contributing factor to both diseases.12,13 However, to our knowledge, no studies published to date have comprehensively explored the potential common genetic etiology of schizophrenia and psoriasis.
In order to avoid false-positive findings in GWAS, a stringent significance threshold is usually applied. When sample sizes are small, conventional GWAS of complex diseases may overlook variants with small or moderate effects. Several complementary approaches have been developed to improve the characterization of the genetic basis of common disorders and traits in GWAS settings (e.g., gene-based association tests, gene-set analyses).14,15 In addition, it is now recognized that many common disorders and traits have a polygenic basis (i.e., many genes, each with a small effect, contribute to a phenotype) that can be characterized through the use of whole genome genotype data. In fact, the genetic architecture for many common diseases and complex traits has been characterized through polygenic methods.16–18 Given that most common disorders and traits have been shown to have a polygenic basis, many researchers have explored the possibility that many complex disorders and traits, including schizophrenia and psoriasis, actually share genetic determinants (i.e., are genetically correlated to some degree).19–26 This trend appears to be even more pronounced in diseases of autoimmune origin.27 In this light, we tested our hypothesis of a shared genetic basis of schizophrenia and psoriasis using large cohorts previously studied in independent GWAS.
Methods
Study design, samples and quality control
We pursued our analyses in 3 stages. In stage 1, we used the mixed linear model to explore the polygenic architecture of schizophrenia and psoriasis in isolation and then combined the data associated with each and reran the analysis. In stage 2 we used 2 popular methods to quantify the possible genetic correlation between schizophrenia and psoriasis: bivariate mixed linear modelling, as implemented in the genome-wide complex trait analysis (GCTA) software suite, and polygenic scoring profile using standard methods. Finally, in stage 3 we used standard single variant association tests to identify candidate variants that might be associated with both conditions in a more pronounced way than others, and we looked for common molecular pathways affected by the most strongly associated variants using gene-set enrichment analyses. This 3-stage strategy is summarized in Appendix 1, Fig. S1, available at jpn.ca.
Two previously reported independent schizophrenia and psoriasis cohorts of Han Chinese descent were used in the present study.3,4 The schizophrenia diagnosis was made by at least 2 experienced psychiatrists according to DSM-IV criteria. No participants had severe medical complications or other psychiatric disorders. The clinical diagnosis of psoriasis was also confirmed by at least 2 dermatologists. To enrich for individuals with a likely overt genetic basis for their condition, we preferentially selected individuals with type 1 psoriasis (early age of onset) who had a family history of disease. All control individuals were clinically determined to be free of autoimmune or psychiatric disorders, psoriasis or family history of such disorders (including first-, second- and third-degree relatives). We collected additional clinical and demographic information on all cohorts through questionnaires. Individuals were categorized into 3 groups based on disease status: a psoriasis case–control cohort, a schizophrenia case–control cohort and a combined cohort (i.e., all individuals from both studies). There were no individuals common to both the schizophrenia and psoriasis cohorts. It should be noted that we did not make the diagnosis and exclusion of schizophrenia in the psoriasis cohort, and vice versa. All participants provided written informed consent. This study was approved by the institutional review boards of The First Affiliated Hospital, Anhui Medical University and Institute of Mental Health, Peking University.
Genotyping was performed on the Illumina Human 610-Quad BeadChip according to the manufacturer’s specifications. We applied quality control procedures, resulting in the exclusion of markers with a minor allele frequency (MAF) less than 0.01, call rate less than 0.9, deviation from Hardy–Weinberg equilibrium in the controls (p < 1 × 10−6), all markers on the X, Y and mitochondrial chromosomes and the copy number variant (CNV)–related probes. Individuals with low overall call rates (< 0.9) were excluded from the study. We performed genome-wide genotype imputation on autosomal chromosomes using IMPUTE2 software and the 1000 Genomes Project phase 1 integrated variant set reference panel.28 Imputed calls were made using a threshold of 0.9 in the software GTOOL (www.well.ox.ac.uk/~cfreeman/software/gwas/gtool.html). Markers with info values less than 0.5 were excluded. Imputed markers were also subject to the quality control procedures implemented on the genotyped data described above.
Estimation of the phenotypic variation explained by polygenic factors
We computed genetic relatedness matrices (GRM) in each of the 3 cohorts (schizophrenia, psoriasis, combined) from genotyped and imputed autosomal markers using the GCTA software.16 We excluded 1 individual from each pair of related individuals (relatedness < 0.025). This resulted in the exclusion of 11, 14, and 67 individuals in the psoriasis, schizophrenia and combined cohorts, respectively. Restricted maximum likelihood (REML) estimation was applied to estimate the proportion of variation explained in the liability of a disease by genetic markers (h2SNP) in each of the cohorts. To facilitate the analyses, we set the prevalence of psoriasis, schizophrenia, and the combined cohort to 0.47%, 1%, and 1.47%, respectively, based on published data.5,29 We included the first 20 principal components from the GRM as covariates in the analyses to accommodate genetic background heterogeneity and potential stratification. The contribution of genetic markers to disease status in each of the cohorts was further broken down into components based on chromosome and MAF. In the former case, genetic markers were partitioned into groups based on chromosome. We estimated GRMs for each chromosome, and the proportion of disease liability explained by each chromosome was calculated independently (separate analysis) as well as jointly with all other chromosomes (joint analysis). Likewise, genetic markers were partitioned based on MAF (uncommon: 0.01 < MAF < 0.05; common: MAF > 0.05), and independent and joint analyses were performed.30 We conducted an REML analysis using the GCTA package to characterize the polygenic architecture of schizophrenia, psoriasis and the combined phenotype.16 To quantify the genetic correlation (or coheritability) between schizophrenia and psoriasis, we implemented a bivariate extension of the REML methods in the GCTA software with the combined cohort.31
Polygenic scoring profile
The shared contribution of genetic variants including, but not limited to, the HLA region, was assessed using markers genotyped in both the schizophrenia and psoriasis cohorts. Markers were pruned based on linkage disequilibrium (LD) with neighbouring markers (maximum threshold for pairwise r2 of 0.25, 200–single nucleotide polymorphism [SNP] window). We then treated the schizophrenia data set as the discovery (i.e., training) set and the psoriasis data set as the target (i.e., test) set, and vice versa. Based on the association statistics in the training set, we selected 5 sets of markers using various significance thresholds (pT; markers with pT < 0.001, pT < 0.05, pT < 0.1, pT < 0.5 as well as all markers). In the target set, we calculated genetic risk scores for each individual as the sum of the risk alleles weighted by the corresponding natural log of odds ratios estimated in the discovery set. The scoring was implemented in the PLINK tool set via the score function.32 A logistic regression model was then fit using the estimated genetic risk score as the independent variable and disease as the dependent variable. We calculated the area under the receiver operating characteristic (ROC) curve (AUC) to assess the degree to which schizophrenia risk estimates predicted susceptibility to psoriasis and vice versa.
Genome-wide association analysis and modelling in the HLA region
Single variant association analyses were pursued assuming the additive allelic model as implemented in the PLINK 1.07 software suite for each of the 3 cohorts.32 The first 20 principal components extracted from a GRM across all individuals in a study were included as covariates to account for possible genetic stratification in the sample. After LD-based pruning, markers in the HLA region that exhibited a pronounced association with disease in the combined cohort (p < 1 × 10−5) showed nominal association with both the schizophrenia and psoriasis cohort (p < 0.05), and displayed the same direction of effect and were therefore included in specific additional analyses (referred to as “marker subset A”).
All 680 genotyped and imputed markers in the HLA region were considered in a least absolute shrinkage and selection operator (LASSO) logistic regression model in the combined cohort via the glmnet analysis package in R software version 3.0.33 We performed 10-fold cross-validation to select the value of the tuning parameter λ, which maximized the AUC (Appendix 1, Fig. S2). Markers with nonzero effect estimates from the LASSO model were noted (referred to as “marker subset B”). Twelve markers were common in both marker subsets A and B. These markers were then fit in a stepwise logistic regression model in the combined cohort, and parameter estimates and corresponding conditional association statistics were recorded. The results of this analysis suggested that 7 of the 12 markers were not associated with disease when the effects of the other markers were considered in a single all-encompassing model. Using the remaining 5 independently associated markers, we calculated a genetic risk score for each individual in the combined cohort using a weighted allele counting approach29 in which weights were assigned based on the effect size estimates obtained from the logistic regression model. Bins were constructed based on the risk score quartiles among controls, and odds ratios in each bin were calculated with respect to the first quartile.
Functional annotation and gene ontology
The functional effects of the 12 HLA markers identified above as well as the non-HLA markers with suggestive significance in the combined cohort (p < 1 × 10−5; n = 12 markers) were investigated with information from the ENCODE database using HaploReg.34 The overlap in shared biological processes between psoriasis and schizophrenia was explored using gene set enrichment analysis (GSEA) implemented in the Aligator software package.15 We performed the GSEA analyses in the schizophrenia, psoriasis and combined cohorts individually. Only markers exceeding a single SNP association threshold (p < 0.001) in their respective data sets were included.
Independent validation
Findings were validated from the summary statistics of 2 GWAS in schizophrenia35 (1630 cases and 1860 controls) and psoriasis36 (441 cases and 2420 controls) of Singapore Chinese descent. The schizophrenia data set came from the Singapore Translational and Clinical Research in Psychosis (STCRP) Study. All patients were recruited from the Institute of Mental Health in Singapore from 2005 to 2008 and were aged 18–83 years. The diagnosis of schizophrenia was made using the Structured Clinical Interview for DSM-IV, Research Version, Patient Edition (SCID-I/P). The controls were from the Singapore Prospective Study Program 11, who were randomly sampled from the Singapore population. All these samples were genotyped by Illumina 1M duo3 array. The Singapore psoriasis cohort included individuals who were genotyped using the Illumina Human550 BeadChip, whereas the controls were genotyped using Illumina Human550, Illumina Human610 Quad and 1M duo3 BeadChip. There was no overlap between the schizophrenia and psoriasis validation cohorts. Genetic variant imputation was performed individually in each study using the 1000 Genomes Project reference panel, and unified quality controls (impute INFO > 0.8, MAF > 0.01, Hardy–Weinberg equilibrium test p > 1.00 × 10−5 in controls, dosage threshold > 0.9, call rate > 0.99), and LD-pruning criteria (SNPs pruned by indep-pairwise 50 5 0.3 parameter in PLINK 1.07) were applied. We considered only biallelic markers in the analyses. The probabilistic significance of variants exhibiting consistent direction of effect between the validation studies was assessed using a binomial test, assessing Pr(X > x), where X~bin(n,p), n = the number of markers reaching a particular significance level in both studies, p = 0.5, and x = the number of markers at the corresponding significance level with the same direction of effect.
Results
Population
Individuals were categorized into 3 groups based on disease status: a psoriasis case–control cohort (1139 cases and 1132 controls), a schizophrenia case–control cohort (744 cases and 546 controls) and a combined cohort (1883 cases and 1678 controls). There was no overlap between the schizophrenia and psoriasis cohorts. We excluded 79 individuals with low overall call rates (< 0.9) from our analysis.
Summary description
After genomic imputation and quality control, we obtained genetic data on roughly 5.6 million high-quality autosomal SNPs in the schizophrenia, psoriasis and combined cohorts. Inspection of the single marker association results in the combined cohort revealed no obvious bias or population stratification issues (genomic inflation factor λGC = 1.01; Appendix 1, Fig. S3). The quantile-quantile (QQ) plot and Manhattan plot in the combined cohort are shown in Appendix 1 (Fig. S4 and Fig. S5). At the upper tail of the distribution, the QQ plot showed an obvious deviation from the null distribution owing to the effects of the HLA region. Thus, after omitting SNPs in this region (chr.6: 29 700 kb– 33 300 kb), the distribution of observed p values largely fit the null (i.e., uniform) distribution. In view of evidence of a shared common polygenic contribution, the deviation in the QQ plot after omitting SNPs in the HLA region potentially shows the aggregate effects of a moderate number of low-risk variants.
Proportion of liability explained by common variants in schizophrenia and psoriasis
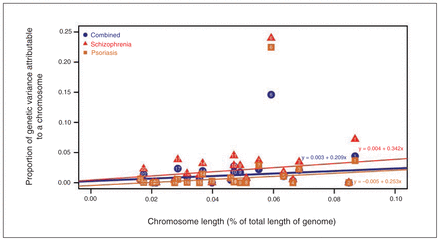
We discovered that 41.3% ± 13.9% (p = 0.002) of the variation in genetic liability for schizophrenia was captured by genotyped and imputed variants. The proportion of liability explained by markers on each chromosome was linearly correlated with the length of the respective chromosome (R2 = 0.1, p = 0.09; Fig. 1 and Appendix 1, Fig. S6). A partition of markers based on MAF (common: MAF > 0.05; uncommon: 0.01 < MAF < 0.05) suggested that common variants were the main contributors to the polygenic liability (> 85%; Appendix 1, Fig. S7 and Table S1). Meanwhile, we found that 34.9% ± 6.0% (p = 9.0 × 10−9) of the variation in genetic liability for psoriasis was captured by genotyped and imputed variants.37 We observed that chromosome 6 was the main contributor to SNP heritability (h2SNP) in both the schizophrenia and psoriasis cohorts (Fig. 1 and Appendix 1, Fig. S6). Furthermore, the genetic variance explained by each chromosome was highly correlated between schizophrenia and psoriasis (R2 = 0.93, p = 0.005).
The distribution of genetic variance explained by each autosomal chromosome in a joint analysis in 3 cohorts. The X axis denotes the length proportion of each individual chromosome in the whole genome. The Y axis denotes the proportion of total genetic variation. The results in the schizophrenia, psoriasis and combined cohorts are depicted in red triangles, brown squares and blue circles, respectively. The numbers in the circles/squares/triangles are the chromosome numbers. The linear correlation between the chromosome length proportion and the proportion of total genetic variation is described using 3 equations in different colors (red: schizophrenia; orange: psoriasis; blue: combined phenotype).
Coheritability and shared genetic etiology of schizophrenia and psoriasis
We next evaluated SNP heritability in the combined cohort (n = 3494). We found that 29.4% ± 5.0% (p = 2.0 × 10−8; Appendix 1, Table S2) of the genetic liability for disease was captured by genotyped and imputed markers. We assessed genome-wide pleiotropy between schizophrenia and psoriasis using marker-based coheritability (Appendix 1, Table S3). We estimated the coheritability (rg SNP) as 0.21 ± 0.20, (p = 0.10). This finding provides preliminary evidence that schizophrenia and psoriasis possibly share a common genetic etiology. Our analysis of each chromosome individually suggested that the coheritability was distributed roughly equally across chromosomes, suggesting in turn that a shared pattern of polygenic contribution to disease exists (Appendix 1, Table S4).
Encouraged by previous results from GWAS initiatives in schizophrenia and psoriasis that have shown susceptibility variants in the HLA region (as well as our discoveries on chromosome 6), we extracted all 49 332 SNPs in an extended HLA region (chr.6: 25–34 MB) and found that 7.0% ± 1.0% (p < 1 × 10−8) of genetic liability for disease was captured by the HLA variants in the combined cohort (Appendix 1, Table S5). We then estimated the contribution of each individual chromosome to h2SNP in the combined cohort. We again found that the variance in disease liability explained by each chromosome was linearly correlated with its length when chromosomes were considered independently (R2 = 0.42, p = 0.001) after omitting chromosome 6 owing to its substantial contribution (Fig. 1). However, the correlation was considerably smaller when examining the contribution of each chromosome, conditional on the others (R2 = 0.10, p = 0.09; Appendix 1, Fig. S6). Finally, we again partitioned markers into 2 categories based on MAF. We found that roughly 80% of the estimated heritability of disease was due to common variants (Appendix 1, Fig. S7 and Table S6).
Polygenic scoring profile analysis
After pruning based on LD, we identified 222 161 independent SNPs genotyped in both the schizophrenia and psoriasis cohorts. We constructed a polygenic risk score using data from the schizophrenia cohort and tested this model in the psoriasis cohort (and vice versa) using 5 different p value thresholds. We discovered that genetic risk in 1 cohort was a significant predictor of genetic risk in the alternative cohort (Table 1). Interestingly, the AUC was highest when using more moderate significance thresholds in the discovery set, implying that genetic background and/or variants with small effects may contribute to the shared etiology between schizophrenia and psoriasis.
The results of polygenic scoring profile analysis
Association analysis and modelling in the HLA region
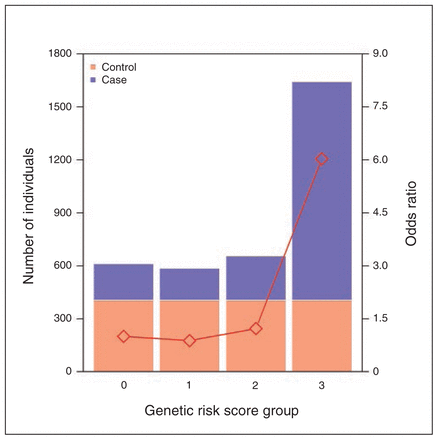
After LD pruning, we identified 565 SNPs with nominal association (p < 0.05) in both the schizophrenia and psoriasis cohorts. More of these markers (55%) displayed the same direction of effect that would be expected by chance (binomial test p = 0.02). Many of these markers (n = 22) were found in the HLA region and showed some evidence for association (p < 1 × 10−5) in the combined cohort (subset A). We then modelled all 680 markers in the HLA region with nominal significance (p < 0.05) in the schizophrenia, psoriasis and combined cohorts simultaneously using a LASSO penalized regression model. Markers with nonzero regression coefficients were recorded (subset B). Twelve markers reached our criteria for subset A and subset B (Table 2, Appendix 2, Table S9 and S10). Among those, we performed forward and backward stepwise regression and discovered 5 SNPs that jointly contributed to disease risk in the combined cohort (Appendix 1, Fig. S8 and Appendix 2, Table S9 and S10). In construction of a genetic risk score based on these 5 markers, we found that the highest-risk group was 6 times more likely than the lowest-risk group to have disease (p < 2.00 × 10−16; Fig. 2).
The combined risk effect distribution of 5 single nucleotide polymorphisms (SNPs) in the human leukocyte antigen (HLA) region in the combined cohort. The X axis denotes the genetic risk score group. The left Y axis denotes the number of individuals in each group. The right Y axis represents the odds ratio. Group 0 = the genetic risk score is less than the 25% quartile of that in controls. Group 1 = the individual score is less than the median but larger than the 25% quartile in controls. Group 2 = the genetic risk score is larger than the median but less than the 75% quartile of that in controls. Group 3 = the score is larger than the 75% quartile of that in controls. Odds ratios were calculated with respect to Group 0. The number of controls and cases are in orange and blue, respectively.
The association results of 12 SNPs in the HLA region implicated in LASSO model
Gene ontology analysis
To explore potential shared biological processes between schizophrenia and psoriasis, we implemented gene ontology analyses in the combined, schizophrenia and psoriasis cohorts separately. We found 18 categories showing significant enrichment in each of the 3 data sets (Appendix 2, Table S8). Most were involved in antigen process and presentation and in endoplasmic reticulum.
Independent validation in the Singapore Chinese population
After genomic imputation and LD pruning, we found that 68 922 SNPs from approximately 6 million genotyped and imputed bi-allelic markers were shared in the Singapore schizophrenia and psoriasis data sets. We used these SNPs to assess the consistency of the p values for association between schizophrenia and psoriasis. Markers were placed into bins based on their significance level obtained in both data sets: p ≤ 0.01, 0.03, 0.05, 0.07, 0.08, 0.09 and 0.10. Then the number of markers with the same direction of effect in each bin was enumerated, and the consistency of directional effect was evaluated using a binomial test (Appendix 1, Table S11). The results suggest that there are indeed genetic variants that exhibit pleiotropic effect in the sense that they are associated with the risk for both schizophrenia and psoriasis, although the pleiotropic effect of any single variant may be small. In addition, although the independent validation data sets didn’t provide direct validation to our findings in the HLA region, single locus analyses of these independent validation data sets, suggested that several SNPs residing in the HLA region conferred significant risk for both schizophrenia and psoriasis, and the effect of the deleterious SNP allele was in the same direction in both conditions, adding further evidence to our initial findings involving the HLA region (Appendix 1, Table S12).
Discussion
Main findings and implications
In the present study, we characterized the polygenic architecture of schizophrenia and psoriasis and explored the possibility of shared genetic etiology between schizophrenia and psoriasis in the Han Chinese population. Our study provides preliminary evidence to support the polygenic basis of both diseases. We found evidence that a shared genetic etiology (i.e., genetic correlation) exists between schizophrenia and psoriasis, and we were able to quantify, to our knowledge for the first time, the strength of the genetic correlation (or coheritability) through the use of whole genome genotype array data. In particular, the contribution of the HLA region appears to be shared between diseases, and we identified 5 SNPs residing in the HLA region contributing in a more pronounced way than other variants to the risk of both diseases. These results highlight the potentially important role of antigen presentation and endoplasmic reticulum pathways in the genetically mediated etiology of schizophrenia and psoriasis.
Ultimately, we explored the polygenic architecture of schizophrenia and psoriasis in the Han Chinese samples. The finding verified the polygenic characteristics of schizophrenia in our samples.30 In addition, our results highlight the important role of common variants in the etiology of both schizophrenia and psoriasis. In general, it appears that both diseases follow a polygenic model of inheritance30,37 (i.e., beyond the HLA region), predominantly influenced by common variants with small effects. It has been shown that 23% of the variation in genetic liability for schizophrenia is captured by common variants in the European population.30 The difference between our estimate of variation in liability for schizophrenia (41.3%) and liability in European samples is possibly owing to sampling error given the different sample sizes used in the studies. Furthermore, we tested the hypothesis of a shared genetic etiology for schizophrenia and psoriasis using single-marker, multimarker and gene ontology analyses with 2 independent Han Chinese GWAS cohorts. We identified evidence of genetic correlation or coheritability between schizophrenia and psoriasis across several analytical approaches. Moreover, our independent validation in the Singapore Chinese provided complementary evidence of this common genetic etiology. Recently, evidence has been found for genetic pleiotropy between many complex human diseases, especially immune-mediated disorders.19,21,22,24–26,29 Schizophrenia has been shown to share a common genetic basis not only with other psychiatric diseases (e.g.. bipolar disorder), but also with metabolic (e.g., type 2 diabetes) and immune disorders (e.g., rheumatoid arthritis).19,25,26 We estimated that 21% of the genomic contribution to both schizophrenia and psoriasis could be attributed to the shared genetic variants. This finding was validated using a polygenic scoring profile analysis and replicated in our independent Singapore data sets. Clinical cases of individuals with both schizophrenia and psoriasis have been documented.6 Large epidemiological studies have shown that the higher incidence of psoriasis in patients with schizophrenia and even in individuals with a family history of schizophrenia in both Chinese and European populations.7–10,20 While we demonstrated a shared genetic etiology between these diseases, common environmental factors may still play a role in the disease coetiologies (e.g., stress event in epidemiological studies).38,39 The additional role of environmental factors and how such environmental factors may influence genetic factors (i.e., gene × environment interactions) remains to be explored. As we used genome-wide genotype array data, we could at least argue that our study is not likely to be attributable to shared environmental factors because it is highly unlikely that environmental stressors or stimuli correlate perfectly with, for example, a genome-wide risk score for each disorder. In addition, our result demonstrating coheritability and consistent effective direction of common variants between schizophrenia and psoriasis agrees with clinical observation and prior epidemiological evidence. Ultimately, we argue that the common genetic etiology would account for the comorbidity of schizophrenia and psoriasis in proportion. However, considering the relatively moderate sample size of our study, further efforts to more accurately estimate coheritability should be explored.
It is strongly believed that the immune process plays an important role in schizophrenia and psoriasis.2,3,11 Recent studies provide evidence that susceptibility to both diseases can be attributed to the HLA variants.1,3,4,29,40 In this study, we highlighted the important role of chromosome 6, and investigated markers residing in the HLA region. By modelling markers in this region, we identified 5 SNPs that jointly contribute to disease susceptibility. Our results confirm earlier reports that suggest the HLA region is involved in schizophrenia and psoriasis1,3,4,29,40 and shed light on their polygenic architecture. The 5 SNPs we assessed in the HLA region that jointly contribute to disease may contribute to the etiology of schizophrenia and psoriasis through a regulatory effect (Appendix 1, Table S9). The markers we identified implicate HLA-B, HLA-C, MICB and HLA-DRB as joint contributors to disease susceptibility. Notably, 1 of these markers (rs41293883) is a missense polymorphism in the MICB gene that results in amino acid change from threonine to isoleucine, although it is predicted to be benign in Polyphen2. We did not impute the HLA molecules or amino acid status. Owing to the complexity of the HLA region and the need for more focused analysis involving the HLA region, future studies are needed.
Beyond the antigen presentation processing pathways, we found evidence that several biological pathways related to the endoplasmic reticulum harbour variants that are shared between schizophrenia and psoriasis. The endoplasmic reticulum plays an important role in the transportation of calcium ion in cells.41 Our findings therefore provide some level of validation of the potential role of calcium signalling in the etiology of schizophrenia and psoriasis.13,42,43
Strengths and limitations
The strength of our study is rooted in the use of multiple complementary statistical approaches designed to identify genetic correlations that all suggest a shared genetic etiology between schizophrenia and psoriasis. We were able to quantify the genetic correlation (coheritability) between schizophrenia and psoriasis through genome-wide array data, and to our knowledge this had not been shown before. In addition, we were able to replicate our results in an independent Singapore Chinese population.
Our study’s limitations revolve around our relatively small samples, which led to a standard error on our estimates of genetic correlation and coheritability from the bivariate linear mixed model. In addition, the AUC value obtained from our polygenic scoring profile analyses is not very large. Larger studies should be pursued to replicate and validate our findings. Based on previous findings suggesting that variants in the HLA region are most likely to be associated with both diseases,3,29 we tested the hypothesis that specific variants in the HLA region could predispose to both diseases. We found credible evidence for this phenomenon. However, the complex LD structure among variants in the HLA region complicates the prioritization of specific variants, and as such our finding that 5 HLA variants exhibit associations warrants validation in independent samples. In addition, the precise molecular mechanism underlying these association signals remains to be elucidated.
Conclusion
We identified a polygenic contribution to schizophrenia and psoriasis in the Han Chinese population. We discovered a shared genetic etiology between the disorders, a large fraction of which could be accounted for mainly by common variants, particularly in the HLA region. The mechanism for this shared genetic basis seems to involve immune and calcium-signalling pathways.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81370044, 81000692, 81273301, 81072461, 81130031, 81222022 and 81222017) and China Council of Scholarship (201208340003), Youth Project of the Outstanding Talents of Organization Department of the CPC Central Committee Program (31200939), the Pre-National Basic Research Program of China (973 Plan) (2012CB722404), Youth Project of Anhui Province Natural Science Foundation (1208085QH145), Anhui High Education Young Talent (X. Yin) and Anhui Medical University Ph.D. Fund (XJ201429). NEW is supported by the NIH grant 1UL1TR001114. NJS and his lab are supported by NIH grants U19 AG023122-09, R01 DA030976-05, R01 MH094483-03, R01 AG035020-05, R01 MH100351-02, R21 AG045789-01A1 as well as grants from Human Longevity, Inc., Johnson and Johnson, the Tanner Foundation, and the Stand-Up-to-Cancer organization. The Singapore Translational and Clinical Research in Psychosis is supported by the National Research Foundation Singapore under the National Medical Research Council Translational and Clinical Research Flagship Program (Grant No.: NMRC/TCR/003/2008). We would like to thank all the individuals participating in our study, Siow Ann Chong, PhD in Institute of Mental Health Singapore, as well as other researcher and physicians who helped to recruit samples and collect information.
Footnotes
Competing interests: None declared.
Contributors: X. Yin, N.J. Schork and X. Zhang designed the study. X. Yin, K. Wang, W. Yue, N. Norgren, W. Yao, X. Jiang, B. Wu, Y. Cui, C. Shen, H. Cheng, F. Zhou, G. Chen, X. Zuo, X. Zheng, X. Fan, H. Wang, L. Wang, J. Lee, M. Lam, S. Tai, Z. Zhang, Q. Huang, L. Sun, J. Xu, S. Yang, K. Wilhelmsen and J. Liu acquired the data, which X. Yin, N. Wineinger, L. Wang and N. Schork analyzed. X. Yin, N. Wineinger and N. Schork wrote the article, which all authors reviewed and approved for publication.
- Received June 3, 2015.
- Revision received October 26, 2015.
- Revision received December 15, 2015.
- Accepted December 21, 2015.